Abstract
Introduction: Multiple studies have demonstrated that diffuse large B-cell lymphoma (DLBCL) can be divided into subgroups based on their biology. However, these biological subgroups overlap clinically. While R-CHOP (rituximab, cyclophosphamide, doxorubicin, vincristine, and prednisone) remains the standard of care for treating patients with DLBCL, predicting which patients will not benefit from such therapy is important so that alternative therapy or clinical trials can be considered. Most of the studies stratifying patients select biomarkers first, then explore how these biomarkers can stratify patients based on outcome. We explored the potential of using machine learning to first group patients with DLBCL based on survival, then isolating the biomarkers necessary for predicting these survival subgroups.
Methods: RNA was extracted from tissue paraffin blocks from 379 R-CHOP treated patients with de novo DLBCL, and from 247 patients with extranodal DLBCL. A targeted hybrid capture RNA panel of 1408 genes was used for next generation sequencing (NGS). Sequencing was performed using an Illumina NextSeq 550 System platform. Ten million reads per sample in a single run were required, and the read length was 2 × 150 bp. An expression profile was generated from the sequencing coverage profile of each individual sample using Cufflinks. A machine learning system was developed to classify patients into four groups based on their overall survival. This machine learning approach based on Naïve Bayesian algorithm was also used to discover the relevant subset of genes with which to classify patients into each of the four survival groups. To eliminate the underflow problem commonly associated with the standard Naïve Bayesian classifiers, we applied Geometric Mean Naïve Bayesian (GMNB) as the classifier to predict the survival group for each patient.
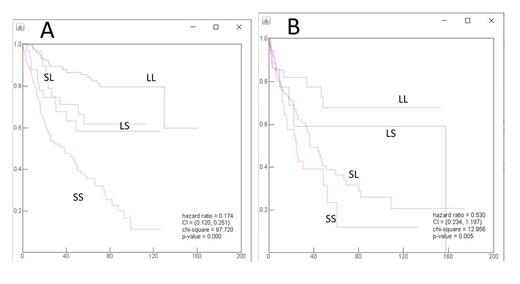
Results: Using machine learning, patients were first divided into two groups: short survival (S) and long survival (L). To refine this model, we used the same approach and divided the patients in each group into two subgroups, generating four groups: long survival in the long group (LL), short survival in the long group (LS), long survival in the short group (SL), and short survival in the short group (SS). The hazard ratio for this model was 0.174 (confidence interval: 0.120-0.251), and P-value <0.0001. After defining these four groups, a machine learning algorithm was used to discover the biomarkers from the expression data of the 1408 genes from NGS data. To reduce the effects of noise and avoid overfitting, we employed a 12-step cross validation to obtain a robust measure. For an individual gene, a generalized Naïve Bayesian classifier was constructed on the training of one of the 12 subsets and tested on the other 11 testing subsets. This allowed us to limit the prediction process to 60 genes for each separation step. Using the selected biomarkers, we classified the patients in the original set (379 patients) into LL, LS, SL, and SS groups and then evaluated the survival pattern of these groups. As shown in Fig. 1A, the selected biomarkers predicted survival as expected in the overall survival groups prior to biomarker selection. For additional validation of the system, we used the selected biomarkers to classify a completely new set of 247 samples of patients with extranodal DLBCL. As shown in Fig. 1B, these selected biomarkers successfully predicted the overall survival in this group of patients with an HR of 0.530 (confidence interval: 0.234-1.197, P=0.005). This classification correlated with cell of origin classification, TP53 mutation status, MYC expression, and IRF4 expression. However, in a multivariate analysis, only TP53 mutation was independent in predicting prognosis (P=0.005) and age (below or over 60) (P=0.01) along with the survival grouping (P<0.000001).
Conclusions: Using a novel machine learning approach with the expression levels of 180 genes, we developed a model that can reliably stratify patients with DLBCL treated with R-CHOP into four survival subgroups. This model can be used to identify patients who may not respond well to R-CHOP to be considered for alternative therapy and clinical trials.
Hsi: AbbVie Inc, Eli Lilly: Research Funding. Ferreri: Ospedale San Raffaele srl: Patents & Royalties; BMS: Research Funding; Pfizer: Research Funding; Beigene: Research Funding; Hutchison Medipharma: Research Funding; Amgen: Research Funding; Genmab: Research Funding; ADC Therapeutics: Research Funding; Gilead: Membership on an entity's Board of Directors or advisory committees, Research Funding; Novartis: Membership on an entity's Board of Directors or advisory committees, Research Funding; Roche: Membership on an entity's Board of Directors or advisory committees, Research Funding; PletixaPharm: Membership on an entity's Board of Directors or advisory committees; x Incyte: Membership on an entity's Board of Directors or advisory committees; Adienne: Membership on an entity's Board of Directors or advisory committees. Piris: Millenium/Takeda, EUSA, Jansen, NanoString, Kyowa Kirin, Gilead and Celgene.: Membership on an entity's Board of Directors or advisory committees, Speakers Bureau. Winter: BMS: Other: Husband: Data and Safety Monitoring Board; Actinium Pharma: Consultancy; Janssen: Other: Husband: Consultancy; Agios: Other: Husband: Consultancy; Gilead: Other: Husband: Consultancy; Epizyme: Other: Husband: Data and Safety Monitoring Board; Ariad/Takeda: Other: Husband: Data and Safety Monitoring Board; Merck: Consultancy, Honoraria, Research Funding; Novartis: Other: Husband: Consultancy, Data and Safety Monitoring Board; Karyopharm (Curio Science): Honoraria.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal